Single-cell and spatial transcriptomics reveal aberrant lymphoid developmental programs driving granuloma formation
Supplementary Website
Study overview
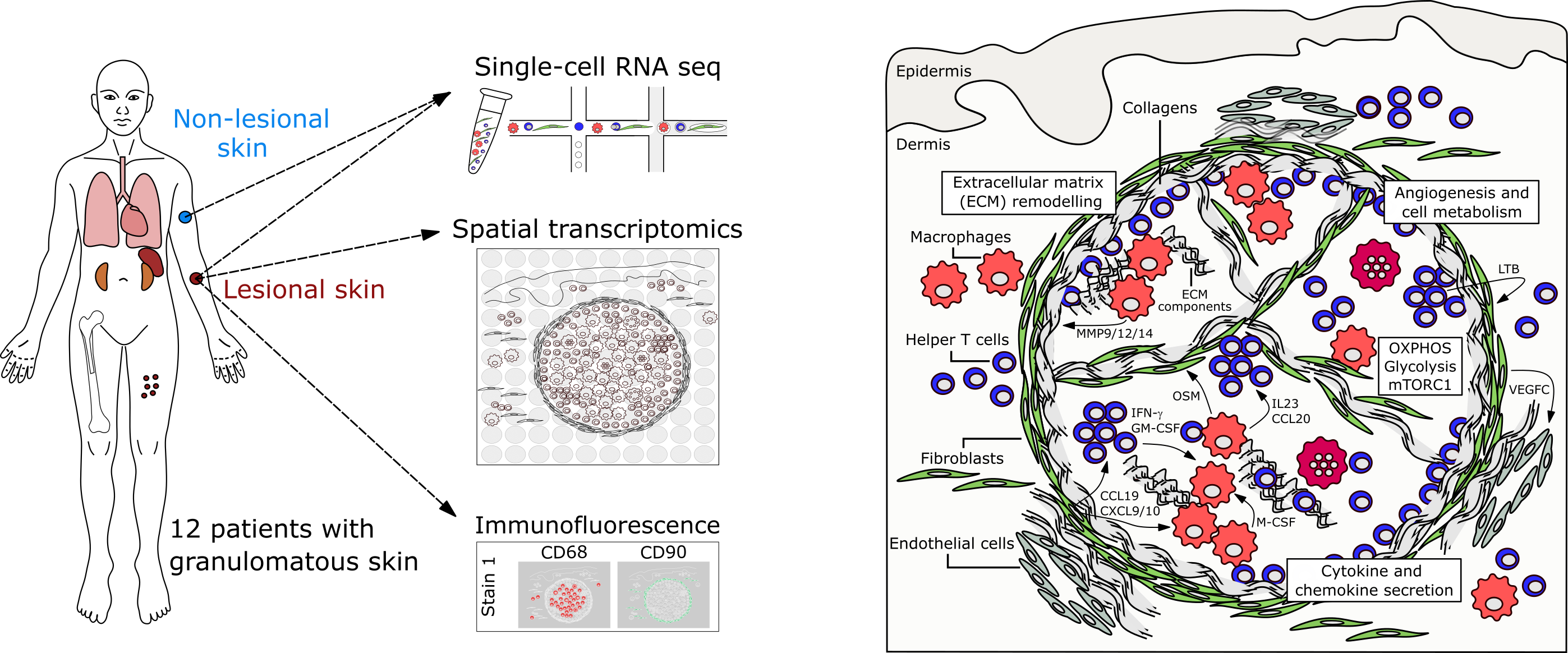
Reconstruction of skin granuloma architecture based on multi-omics data and experimental validation.
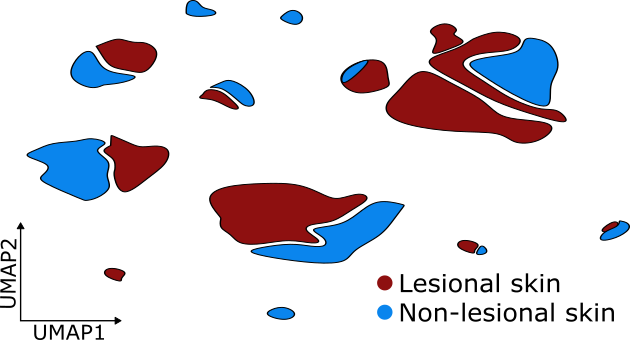
Identification of granuloma-associated cell types
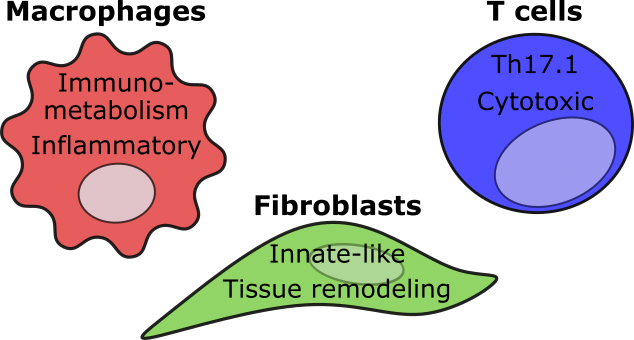
Characterization of disease-driving cell subsets
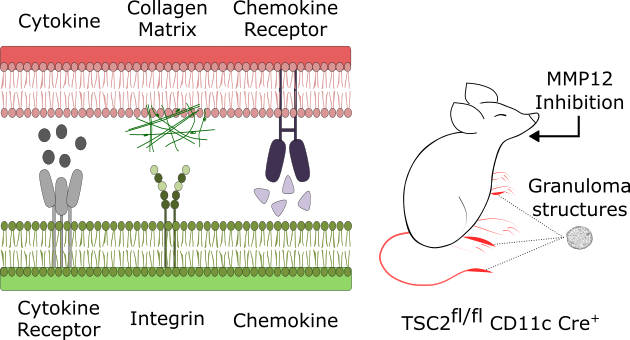
Inference of cellular interaction networks and validation of therapeutic targets
Data download
| Description | Links |
|---|---|
| Raw sequencing data: |
The raw sequencing data are available from the European Genome-phenome Archive repository under accession number EGAS00001006970.
To request access to the raw sequencing data, please e-mail the filled and signed Data Access Agreement to Georg Stary (georg.stary@meduniwien.ac.at). |
| Raw imaging data: |
The microscopy imaging data are available from zenodo: 10.5281/zenodo.7584110. |
| Processed data: |
The processed sequencing data (count matrices) are available from the NCBI Gene Expression Omnibus repository repository under accession number GSE192461. |
| Analysis code: |
The analysis source code is available as a ZIP archive or via zenodo: 10.5281/zenodo.7523056. |
| Supplementary data: |
The projection of literature-derived gene signatures on the spatial data are available as a ZIP archive. |
Citation
If you use these data in your research, please cite:
Thomas Krausgruber*, Anna Redl*, Daniele Barreca*, Konstantin Doberer, Daria Romanovskaia, Lina Dobnikar, Maria Guarini, Luisa Unterluggauer, Lisa Kleissl, Denise Atzmüller, Carolina Mayerhofer, Aglaja Kopf, Simona Saluzzo, Clarice X. Lim, Praveen Rexie, Thomas Weichhart, Christoph Bock#, Georg Stary#
Single-cell and spatial transcriptomics reveal aberrant lymphoid developmental programs driving granuloma formation.
Immunity 2023. DOI: 10.1016/j.immuni.2023.01.014.
* These authors contributed equally to this work
# Correspondence: georg.stary@meduniwien.ac.at (G.S.), cbock@cemm.oeaw.ac.at (C.B.)